Abstract
Background
Genus Adonis L. contain approximately 40 annual and perennial species, which are widely distributed in the temperate zones of Asia and Europe, and less frequently in southwestern Asia, northern Africa and the Mediterranean region. The aim of the study was to evaluate the phylogenetic relationship among Adonis taxa collected from Türkiye based on nrDNA Internal transcribed spacer (ITS) markers.
Methods
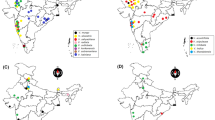
Samples of 64 individual genotypes from 21 populations of 10 Adonis taxa were collected from different regions of the country during vegetation period between 2014 and 2018. ITS1, ITS4, P16 and P25 primers within ITS technique was used to genotype the plant materials. Then, genotypic data was used to estimate magnitude and organization of infraspecific variation in different populations of Adonis.
Results
About 600 bp DNA sequences were obtained from each 64 Adonis genotypes belonging to 21 different populations. The dendrogram obtained from Adonis taxa and out-group sequences had two large main groups. While the out-group species were placed in the first large main group, the sect. Consiligo (perennial) and sect. Adonis (annuals) were placed in different sub-groups of the second large main group. Genetic similarity among Adonis taxa varied between A. microcarpa and A. dentata (98.46%). Principal component analysis indicated that two important components in Adonis taxa genotypes. The expected heterozygosity ranged from 0.0252 (sub-population A) to 0.3460 (sub-population C), with an average of 0.1154. In addition, population differentiation measurements (Fst) ranged from 0.0025 (sub-population C) to 0.9016 (sub-population A) with a relatively high average 0.6601.
Conclusions
Present analyses revealed that phylogenetic classification (grouping) of Adonis taxa largely depended on morphological structure and present ITS primers were quite efficient in putting forth the genetic diversity of such species. The results of this study suggested that ITS markers could be used in the identification of genetic diversity among the Adonis taxa. The results obtained from molecular data can be used to explore the genetic variation pattern, population structure, and the evolutionary history of genus Adonis in the future.






Similar content being viewed by others
References
Jury SL (1978) Ranunculaceae. In: Heywood VH et al (eds) Flowering plants of the world. Mayflower Books, New York
Anderson CL, Bremer K, Friis EM (2005) Dating phylogenetically basal eudicots using rbcL sequences and multiple fossil reference points. Am J Bot 92:1737–1748
Emadzade K, Lehnebach C, Lockhart P, Hörandl E (2010) A molecular phylogeny, morphology and classification of genera of Ranunculeae (Ranunculaceae). Taxon 59:809–828
Guner A, Aslan S, Ekim T, Vural M, Babac MT (2012) Türkiye Bitkileri Listesi (Damarlı Bitkiler). Nezahat Gökyiğit Botanik Bahçesi ve Flora Araştırmaları Derneği, İstanbul
Christenhusz MJM, Byng JW (2016) The number of known plants species in the world and its annual increase. Phytotaxa 261:201–217
Mabberley DJ (1990) The plant book. Cambridge University Press, New York
Tamura M (1990) A new classification of the family Ranunculaceae 1. Acta Phytotaxon Geobot 41:93–101
Wang WT (1994a) Revision of Adonis (Ranunculaceae) I. Bull Bot Res 14:1–31
Wang WT (1994) Revision of Adonis (Ranunculaceae) II. Bull Bot Res 14:105–138
Ghorbani NM, Azizian D, Sheydaei M, Khatamsaz M (2008) Pollen morphology of some Adonis l. species (Ranunculaceae) from Iran. J Bot 14:165–170
Hossein-Pour A, Karahan F, Ilhan E, Ilcim A, Haliloglu K (2019) Genetic structure and diversity of Adonis L. (Ranunculaceae) population collected from Turkey by inter-primer binding site (iPBS) retrotransposon markers. Turk J Bot 17:1899–1911
Karahan F (2018) The revision of the genus Adonis L. (Ranunculaceae) in Turkey. Dissertation, Hatay Mustafa Kemal University.
Davis PH (1967) Adonis L. In: Davis PH (ed) Flora of Turkey and the East Aegean Islands, vol 1. Edinburgh Univ Press, Edinburgh, pp 141–146
Hoffmann MH (1998) Plant Systematics and Evolution Ecogeographical differentiation patterns in Adonis sect. Consiligo (Ranunculaceae). Pl Syst Evol 211:43–56
Kandemir A, Kaptaner Igcı B, Aytac Z, Fisne A (2019) Contributions to the systematics of the genus Adonis L. (Ranunculaceae) in Turkey. Gazi Univ J Sci 32:1105–1111
Steinberg C (1971) Revisione sistematica e distributiva delle « Adonis » annuali in Italia. Webbia 25:299–351
Chan Son D, Ko C (2013) Aggregated achenes and achene morphology of the Korean Adonis L. and its related taxa in East Asia. Korean J Pl Taxon 43:1–7
Erfanzadeh R, Kahnuj SHH, Azarnivand H, Pétillon J (2013) Comparison of soil seed banks of habitats distributed along an altitudinal gradient in northern Iran. Flora Morphol Distrib Funct Ecol Plants 208:312–320
Menemen Y, Uzel F (2016) Düğünçiçeğigiller (Ranunculaceae) familyasına ait bazı türlerin polen morfolojileri üzerine bir çalışma. Bağbahçe Bilim Dergisi 3:11–19
Heyn CC, Pazy B (1989) The annual species of Adonis (Ranunculaceae)-a polyploid complex. Plant Syst Evol 168:181–193
Johansson JT (1999) There large inversions in the chloroplast genomes and one loss of the chloroplast gene rps16 suggest an early evolutionary split in the genus Adonis (Ranunculaceae). Plant Syst Evol 218:133–143
Altay V, Karahan F, Ozturk M, Hakeem KR, Ilhan E, Erayman M (2016) Molecular and ecological investigations on the wild populations of Glycyrrhiza L. taxa distributed in the East Mediterranean Area of Turkey. J Plant Res 129:1021–1032
Ilhan E, Ozgur S, Tuna GS, Eren AH, Karahan F, Tuna M, Erayman M (2017) Nuclear DNA content variation among Glycyrrhiza taxons collected from East Mediterranean. Fresenius Environ Bull 26:3251–3256
Suh Y, Lee J, Lee S, Lee C, Yeau SH, Lee NS (2002) Molecular evidence for the taxonomic identity of Korean Adonis (Ranunculaceae). J Plant Res 115:217–223
Boronnikova S, Kalendar R (2010) Using IRAP markers for analysis of genetic variability in populations of resource and rare species of plants. Russian J Genet 46:36–42
Despres L, Gielly L, Redoutet B, Taberlet P, (2003) Using AFLP to resolve phylogenetic relationships in a morphologically diversified plant species complex when nuclear and chloroplast sequences fail to reveal variability. Mol Phylogenet Evol 27:185–196
Kropf M, Bardy K, Höhn M, Plenk K (2020) Phylogeographical structure and genetic diversity of Adonis vernalis L. (Ranunculaceae) across and beyond the Pannonian region. Flora 262:151497
Cai YF, Li SW, Liu Y, Quan S, Chen M, Xie YF, Jiang HZ, Wei EZ, Yin NW, Wang L, Zhang R (2009) Molecular phylogeny of Ranunculaceae based on internal transcribed spacer sequences. Afr J Biotechnol 8:5215–5224
Ro KE, Keener CS, McPheron BA (1997) Molecular phylogenetic study of the ranunculaceae: utility of the nuclear 26S ribosomal DNA in inferring intrafamilial relationships. Mol Phylogenet Evol 8:117–127
De Candolle AP (1818) Adonis. In: De Candolle AP (ed) Regni vegetabilis systema naturale, vol 1. Treuttel et Wurtz, Paris, pp 220–226
Spach E (1839) Histoire Naturelle Des Végétaux Phanérogames, 7. Librairie encyclopédique de Roret, Paris
Boissier E (1867) Flora orientalis, vol 1. Bâle & Lyon, Geneve
Bobrov EG (1937) Adonis. In: Komarov VL (ed) Flora of the U.S.S.R. Vol: VII. Koeltz Scientific Book, Koenigstein, pp 403–411
Riedl H (1963) Revision der einjährigen Arten von Adonis L. Ann Naturhistorischen Museums Wien 66:51–90
Davis PH, Mill RRTK (1988). In: Davis PH, Mill RR, Tan K (eds) Flora of Turkey and east Aegean Islands. Edinburgh University Press, Edinburgh, pp 177–180
Tutin T, Akeroyd J (1993) Adonis L. In: Tutin TO et al (eds) Flora Europaea, I, 2nd edn. Cambridge University Press, Cambridge, pp 267–269
Guner A, Ozhatay N, Ekim T, Baser KHC (2000) Flora of Turkey and the East Aegean Islands, vol 11. Edinburgh University Press, Edinburgh
Fu D, Robinson OR (2001) Adonis. In: Wu ZY, Raven PH, Hong DY (eds) Flora of China (Caryophyllaccae through Lardizabalaceae), vol 6. Science Press, Beijing, pp 389–391
Zeinalzadehtabrizi H, Hosseinpour A, Aydin M, Haliloglu K (2015) A modified genomic DNA extraction method from leaves of sunflower for PCR based analyzes. J Biodivers Environ Sci 7:222–225
White TJ, Bruns T, Lee SJ, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Sninsky J (eds) PCR protocols: a guide to methods and application. Academic Press, San Diego, pp 315–322
Liden M, Fukuhara T, Axberg T (1995) Phylogeny of Corydalis, ITS and morphology. Plant Syst Phylogen 9:183–188
Popp M, Erixon P, Eggens F, Oxelman B (2005) Origin and evolution of a circumpolar polyploid species complex in Silene (Caryophyllaceae) inferred from low copy nuclear RNA polymerase introns, rDNA, and chloroplast DNA. Syst Bot 30:302–313
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Letunic I, Bork P (2011) Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:475–478
Wang W, Lu AM, Ren Y, Endress ME, Chen ZD (2009) Phylogeny and classification of Ranunculales: evidence from four molecular loci and morphological data. Perspect Plant Ecol Evol Syst 11:81–110
Jabbour F, Renner SS (2012) A phylogeny of Delphinieae (Ranunculaceae) shows that Aconitum is nested within Delphinium and that Late Miocene transitions to long life cycles in the Himalayas and Southwest China coincide with bursts in diversification. Mol Phylogenet Evol 62:928–942
Jan T, Johansson RKJ (1993) Chloroplast DNA variation and phylogeny of the Ranunculaceae. Pl Syst Evol 187:29–49
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics (Oxford, England) 28:2537–2539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000) Association mapping in structured populations. Am J Hum Genet 67:170–181
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Earl DA, Vonholdt BM (2012) Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Zeinalzadeh-Tabrizi H, Sahin E, Haliloglu K (2011) Principal components analysis of some F1 sunflower hybrids at germination and early seedling growth stage. J Agric Atatürk Uni 42:103–109
Fischer M, Husi R, Prati D et al (2000) RAPD variation among and within small and large populations of the rare clonal plant Ranunculus reptans (Ranunculaceae). Am J Bot 87:1128–1137
Mitton JB, Grant MC (1984) Associations among protein heterozygosity, growth rate and developmental homeostasis. Annu Rev Ecol Syst 15:479–499
Bosh M, Simon J, Molero J, Blache C (1996) Reproductuve biology, genetic variation and conservation of the rare endemic dysploid Delphinium bolosii (Ranssunculaceae). Biol Conserv 86:57–66
Acknowledgements
This study was funded by Scientific Research Commission of Hatay Mustafa Kemal University (Project No: 12580). The authors also are grateful to Dr. Z. Aytaç, Dr. A. Kandemir, Dr. F. Güneş and Dr. G. Akgül for helpful comments on the manuscript.
Funding
This study was funded by Scientific Research Commission of Hatay Mustafa Kemal University (Project No: 12580).
Author information
Authors and Affiliations
Contributions
FK and AI conceived and designed the study; FK and AI developed the plant material; FK, EI and AT performed the DNA extraction, analyzed the ITS data; FK and KH statistical analysis; FK, EI and AT drafted the manuscript; FK, EI and AT revised the manuscript. All authors agree and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All samples were collected from commercial landings, and no ethical approval was required.
Consent to participate
Not applicable.
Consent for publication
All the authors agree to submit and publish an article entitled “Phylogenetic relationship among taxa in the genus Adonis L. collected from Türkiye based on nrDNA Internal transcribed spacer (ITS) markers” in Molecular Biology Reports.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Karahan, F., İlçim, A., Türkoğlu, A. et al. Phylogenetic relationship among taxa in the genus Adonis L. collected from Türkiye based on nrDNA internal transcribed spacer (ITS) markers. Mol Biol Rep 49, 7815–7826 (2022). https://doi.org/10.1007/s11033-022-07607-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07607-7