Abstract
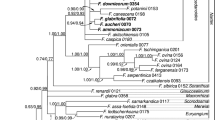
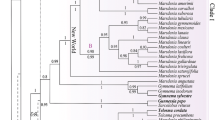
Since the first description of the genus in 1754, the taxonomy of Ananas underwent many fundamental changes and it is still the subject of a vivid debate. We present a phylogeny comprising all seven known Ananas taxa, Pseudananas sagenarius as well as closely related members of Bromelioideae (Aechmea subg. Chevaliera) based on three nuclear markers (agt1, ETS, phyC), five plastid markers (atpB–rbcL, trnL–trnF, matK, two segments of ycf1) and AFLP data. This study reveals a close relationship between Ananas, P. sagenarius, Aechmea tayoensis and Disteganthus basilateralis, and proposes novel relationship of the Ananas clade and Aechmea fernandae. Taxonomic implications of our analysis in particular the recognition of species versus varieties in Ananas are discussed. Furthermore, we could show that the evolution of two traits (scape bracts and the apical coma of the inflorescence) might be interlinked.


Similar content being viewed by others
References
Aguirre-Santoro J (2017) Taxonomy of the Ronnbergia Alliance (Bromeliaceae: Bromelioideae): new combinations, synopsis, and new circumscriptions of Ronnbergia and the resurrected genus Wittmackia. Pl Syst Evol 303:615–640. https://doi.org/10.1007/s00606-017-1394-y
Aradhya MK, Zee F, Manshardt RM (1994) Isozyme variation in cultivated and wild pineapple. Euphytica 79:87–99. https://doi.org/10.1007/BF00023580
Barfuss MHJ (2012) Molecular studies in Bromeliaceae: Implications of plastid and nuclear DNA markers for phylogeny, biogeography, and character evolution with emphasis on a new classification of Tillandsioideae. PhD Thesis, University of Vienna, Vienna
Beadle DA (1998) The Bromeliad cultivar registry. The Bromeliad Society International, Venice
Bollback JP (2006) SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics 7:88. https://doi.org/10.1186/1471-2105-7-88
Bonin A, Bellemain E, Eidesen PB, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics studies. Molec Ecol 13:3261–3273. https://doi.org/10.1111/j.1365-294X.2004.02346.x
Brown GK, Gilmartin AJ (1989) Chromosome numbers in Bromeliaceae. Amer J Bot 76:657–665
Brown GK, Leme EMC (2000) Cladistic analysis in the Nidularioid complex. In: Leme EMC (ed) Nidularium—Bromeliads of the Atlantic Forest. Sexante Artes, Rio de Janeiro, pp 240–247
Butcher D, Gouda EJ (2017) The new bromeliad taxon list. Available at: http://botu07.bio.uu.nl/bcg/taxonList.php. Accessed 27 Jul 2015
Carlier JD, Coppens d’Eeckenbrugge G, Leitao JM (2007) Pineapple genome mapping and molecular breeding in plants. In: Kole C (ed) Fruits and Nuts. Springer, Berlin, pp 331–342
Castello LV, Barfuss MHJ, Till W, Galetto L, Chiapella JO (2016) Disentangling the Tillandsia capillaris complex: phylogenetic relationships and taxa boundaries of Andean populations. Bot J Linn Soc 181:391–414. https://doi.org/10.1111/boj.12400
Collins JL (1936) A frequently mutating gene in the pineapple. Ananas comosus (L.) Merr. Amer Naturalist 70:467–476. Available at:https://doi.org/10.1086/280687
Collins JL (1960) The pineapple, botany, utilization, cultivation. Leonhard Hill, London
Coppens d’Eeckenbrugge G, Duval MF, Van Miegroet F (1993) Fertility and self-incompatibility in the genus Ananas. Acta Hort 334:45–51. https://doi.org/10.17660/ActaHortic.1993.334.4
Coppens d’Eeckenbrugge G, Leal F (2003) Morphology, anatomy and taxonomy. In: Bartholomew DP, Paull RE, Rohrbach KG (eds) The pineapple: botany, production and uses. CAB International, Wallingford, pp 13–32
Degnan JH, Rosenberg NA (2009) Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol 24:332–340. https://doi.org/10.1016/j.tree.2009.01.009
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull Bot Soc Amer 19:11–15
Drábková LZ, Vlček Č (2010) Molecular phylogeny of the genus Luzula DC. (Juncaceae, Monocotyledones) based on plastome and nuclear ribosomal regions: a case of incongruence, incomplete lineage sorting and hybridization. Molec Phylogen Evol 57:536–551. https://doi.org/10.1016/j.ympev.2010.07.022
Duval MF, Coppens d’Eeckenbrugge G (1993) Genetic variability in the genus Ananas. Acta Hortic 334:27–32. https://doi.org/10.17660/ActaHortic.1993.334.2
Duval MF, Noyer JL, Perrier X, Coppens d’Eeckenbrugge G, Hamon P (2001) Molecular diversity in pineapple assessed by RFLP markers. Theor Appl Genet 102:83–90. https://doi.org/10.1007/s001220051621
Duval MF, Buso GSC, Ferreira FR, Noyer JL, Coppens d’Eeckenbrugge G, Hamon P, Ferreira ME (2003) Relationships in Ananas and other related genera using chloroplast DNA restriction site variation. Genome 46:990–1004. https://doi.org/10.1139/g03-074
Felsenstein J (1985) Phylogenies and the comparative method. Amer Naturalist 125:1–15. https://doi.org/10.1086/284325
Fragoso JMV, Huffman JM (2000) Seed-dispersal and seedling recruitment patterns by the last neotropical megafaunal element in Amazonia, the tapir. J Trop Ecol 16:369–385. https://doi.org/10.1017/S0266467400001462
French PA, Brown GK, Bayly MJ (2016) Incongruent patterns of nuclear and chloroplast variation in Correa (Rutaceae): introgression and biogeography in south-eastern Australia. Pl Syst Evol 302:447–468. https://doi.org/10.1007/s00606-016-1277-7
Garcia ML (1988) Etude taxinomique du genre Ananas. Utilisation de la variabilite enzymatique. PhD Thesis, University of Science and Technology, Languedoc, Montpellier
Gelman A, Rubin D (1992) Inference from iterative simulation using multiple sequences. Stat Sci 7:457–472. https://doi.org/10.1214/ss/1177011136
Gitaí J, Paule J, Zizka G, Schulte K, Benko-Iseppon AM (2014) Chromosome numbers and DNA content in Bromeliaceae: additional data and critical review. Bot J Linn Soc 176:349–368. https://doi.org/10.1111/boj.12211
Givnish TJ, Barfuss MHJ, van Ee B, Riina R, Schulte K, Horres R, Gonsiska PA, Jabaily RS, Crayn DM, Smith JAC, Winter K, Brown GK, Evans TM, Holst BK, Luther H, Till W, Zizka G, Berry PE, Systsma KJ (2011) Phylogeny, adaptive radiation, and historical biogeography in Bromeliaceae: insights from an eight-locus plastid phylogeny. Amer J Bot 98:872–895. https://doi.org/10.3732/ajb.1000059
Horres R, Zizka G, Kahl G, Weising K (2000) Molecular phylogenetics of Bromeliaceae: evidence from trnL (UAA) intron sequence of the chloroplast genome. Pl Biol (Stuttgart) 2:306–315. https://doi.org/10.1055/s-2000-3700
Horres R, Schulte K, Weising K, Zizka G (2007) Systematics of Bromelioideae (Bromeliaceae)—evidence from molecular and anatomical studies. Aliso 23:27–43. https://doi.org/10.5642/aliso.20072301.05
Huelsenbeck JP, Nielsen R, Bollback JP (2003) Stochastic mapping of morphological characters. Syst Biol 52:131–158. https://doi.org/10.1080/10635150390192780
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Leal F (1990) On the validity of Ananas monstruosus. J Bromeliad Soc 40:246–249
Leal F, Coppens d’Eeckenbrugge G, Holst BK (1998) Taxonomy of the genera Ananas and Pseudananas—a historical review. Selbyana 19:227–235
Li M, Wunder J, Bissoli G, Scarponi E, Gazzani S, Barbaro E, Saedler H, Varotto C (2008) Development of COS genes as universally amplifiable markers for phylogenetic reconstructions of closely related plant species. Cladistics 24:727–745. https://doi.org/10.1111/j.1096-0031.2008.00207.x
Louzada RB, Schulte K, Wanderley ML, Silvestro D, Zizka G, Barfuss MHJ, Palma-Silva C (2014) Molecular phylogeny of the Brazilian endemic genus Orthophytum (Bromelioideae, Bromeliaceae) and its implications on morphological character evolution. Molec Phylogen Evol 77:54–64. https://doi.org/10.1016/j.ympev.2014.03.007
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop (GCE), San Diego Supercomputer Center, New Orleans, pp 1–8. https://doi.org/10.1109/gce.2010.5676129
Ming R, VanBuren R, Wai CM, Tang H, Schatz MC, Bowers JE, Lyons E, Wang M-L, Biggers E, Zhang J, Huang L, Zhang L, Miao W, Zhang J, Ye Z, Miao C, Lin Z, Wang H, Zhou H, Yim WC, Priest HD, Zheng C, Woodhouse M, Edger PP, Guyot R, Guo H-B, Guo H, Zheng G, Singh R, Sharma A, Min X, Zheng Y, Lee H, Gurtowski J, Sedlazeck FJ, Harkess A, McKain MR, Liao Z, Fang J, Liu J, Zhang X, Zhang Q, Hu W, Qin Y, Wang K, Chen L-Y, Shirley N, Lin Y-R, Liu L-Y, Hernandez AG, Wright CL, Bulone V, Tuskan GA, Heath K, Zee F, Moore PH, Sunkar R, Leebens-Mack J, Mockler T, Bennetzen JL, Freeling M, Sankoff D, Paterson AH, Zhu X, Yang X, Smith JAC, Cushman JC, Paull RE, Yu Q (2015) The pineapple genome and the evolution of CAM photosynthesis. Nat Genet 47:1435–1442. https://doi.org/10.1038/ng.3435
Pelser PB, Kennedy AH, Tepe EJ, Shidler JB, Nordenstam B, Kadereit J, Watson LE (2010) Patterns and causes of incongruence between plastid and nuclear Senecioneae (Asteraceae) phylogenies. Amer J Bot 97:856–873. https://doi.org/10.3732/ajb.0900287
Petit RJ, Excoffier L (2009) Gene flow and species delimitation. Trends Ecol Evol 24:386–393. https://doi.org/10.1016/j.tree.2009.02.011
Posada D (2008) jModelTest: phylogenetic model averaging. Molec Biol Evol 25:1253–1256. https://doi.org/10.1093/molbev/msn083
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of the AIC and Bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808. https://doi.org/10.1080/10635150490522304
R Core Team (2013) R: a language and environment for statistical computing. R Foundation and Statistical Computing, Vienna. Available at: http://www.R-project.org
Revell LJ (2012) Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3:217–223. https://doi.org/10.1111/j.2041-210X.2011.00169.x
Richter W (1978) Zimmerpflanzen von heute und morgen: Bromeliaceen, 4th edn. Neumann Verlag Leipzig, Radebeul
Rogstad SH (1992) Saturated NaCl-CTAB solution as a means of field preservation of leaves for DNA analyses. Taxon 41:701–708. https://doi.org/10.2307/1222395
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Sass C, Specht CD (2010) Phylogenetic estimation of the core Bromelioids with an emphasis on the genus Aechmea (Bromeliaceae). Molec Phylogen Evol 55:559–571. https://doi.org/10.1016/j.ympev.2010.01.005
Schulte K, Horres R, Zizka G (2005) Molecular phylogeny of Bromelioideae and its implications on biogeography and the evolution of CAM in the family (Poales, Bromeliaceae). Senckenberg Biol 85:1–13
Schulte K, Barfuss MH, Zizka G (2009) Phylogeny of Bromelioideae (Bromeliaceae) inferred from nuclear and plastid DNA loci reveals the evolution of the tank habit within the subfamily. Molec Phylogen Evol 51:327–339. https://doi.org/10.1016/j.ympev.2009.02.003
Schulte K, Silvestro D, Kiehlmann E, Vesely S, Novoa P, Zizka G (2010) Detection of recent hybridization between sympatric Chilean Puya species (Bromeliaceae) using AFLP markers and reconstruction of complex relationships. Molec Phylogen Evol 57:1105–1119. https://doi.org/10.1016/j.ympev.2010.09.001
Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molec Biol Evol 16:1114–1116. https://doi.org/10.1093/oxfordjournals.molbev.a026201
Silva BR (2003) Contributions to the understanding of Andean and Amazonian Aechmea subg. Chevaliera (Bromeliaceae). Selbyana 24:46–63
Silvestro D (2013) Diversification in time and space. Methodological advancement and case studies from the Neotropical plant family Bromeliaceae. PhD Thesis, Goethe University Frankfurt, Frankfurt am Main
Silvestro D, Michalak I (2012) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity Evol 12:335–337. https://doi.org/10.1007/s13127-011-0056-0
Silvestro D, Zizka G, Schulte K (2014) Disentangling the effects of key innovations on the diversification of Bromelioideae (Bromeliaceae). Evolution 68:163–175. https://doi.org/10.1111/evo.12236
Siqueira-Filho JA, Leme EMC (2006) Fragments of the Atlantic Forest of Northeast Brazil. Andrea Jakobsson Estúdio, Rio de Janeiro
Smith LB, Downs RJ (1974) Monograph 14, pitcairnioideae (Bromeliaceae). In: Smith LB, Downs RJ (eds) Flora neotropica. Hafner Press, New York
Smith L, Downs RJ (1979) Bromelioideae, Bromeliaceae. New York Botanical Garden, New York, pp 2048–2064
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. https://doi.org/10.1093/bioinformatics/btl446
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucl Acids Res 23:4407–4414. https://doi.org/10.1093/nar/23.21.4407
Wendt T, Canela MBF, de Faria APG, Rios RI (2001) Reproductive biology and natural hybridization between two endemic species of Pitcairnia (Bromeliaceae). Amer J Bot 88:1760–1767
Xu B, Wu N, Gao X-F, Zhang L-B (2012) Analysis of DNA sequences of six chloroplast and nuclear genes suggests incongruence, introgression, and incomplete lineage sorting in the evolution of Lespedeza (Fabaceae). Molec Phylogen Evol 62:346–358. https://doi.org/10.1016/j.ympev.2011.10.007
Zhang J, Liu J, Ming R (2014) Genomic analyses of the CAM plant pineapple. J Exp Bot 65:3395–3404. https://doi.org/10.1093/jxb/eru101
Acknowledgements
We thank Ingo Michalak (University of Leipzig) for his support with the Shimodaira-Hasegawa test. The study was financially supported by the research funding programme “LOEWE—Landesoffensive zur Entwicklung wissenschaftlich-ökologischer Exzellenz” of Hesse’s Ministry of higher education as well as the “Freunde und Förderer” of Goethe-University Frankfurt and Paul Ungerer-Stiftung. We also acknowledge financial support of the German Research Foundation (DFG ZI 557/6-2 and 7-1, SCHU 2426/1-1).
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Mike Thiv.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1.
List of species names included in this study, specimen information, and GenBank accession numbers for all sequences.
Online Resource 2.
Alignment of studied markers (agt1, atpB–rbcL, ETS, matK, phyC, trnL–trnF, ycf1 pos. 1114-2102 and 4550-5532) and AFLP data.
Online Resource 3.
Primer combinations used for selective AFLP amplifications in initial screening and in final analyses.
Online Resource 4.
Results of the Shimodaira-Hasegawa test as implemented in RAxML. Pairwise comparison of single-ML trees inferred by atpB–rbcL, trnL–trnF, matK, ycf1 pos. 1114-2102 and 4550-5532, agt1, ETS, phyC, and AFLP data to test for topological incongruences.
Online Resource 5.
Maximum credibility trees of post-burnin Bayesian analyses, each ran for 30 million generations, based on a. AFLP, b. nuclear (agt1, ETS, phyC) or c. plastid (atpB–rbcL, trnL–trnF, matK, ycf1 pos. 1114-2102 and 4550-5532) data.
Online Resource 6.
Transition model selection for stochastic character mapping and state frequencies per node for Fig. 2. a. The corrected Akaike information criterion (AICc) was used for model selection. b. Table showing the inferred state frequencies calculated across 1000 stochastically mapped character histories (using ER model) for each node in Fig. 2 for two morphological traits: Apical coma (conspicuous, inconspicuous, missing) and Scape bracts (foliaceous, not foliaceous/often imbricate, lacking).
Online Resource 7.
Overview of the published chromosome numbers in the genus Ananas and Pseudananas.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. List of species names included in this study, specimen information, and GenBank accession numbers for all sequences.
Online Resource 2. Alignment of studied markers (agt1, atpB–rbcL, ETS, matK, phyC, trnL–trnF, ycf1 pos. 1114-2102 and 4550-5532) and AFLP data.
Online Resource 3. Primer combinations used for selective AFLP amplifications in initial screening and in final analyses.
Online Resource 4. Results of the Shimodaira-Hasegawa test as implemented in RAxML. Pairwise comparison of single-ML trees inferred by atpB–rbcL, trnL–trnF, matK, ycf1 pos. 1114-2102 and 4550-5532, agt1, ETS, phyC, and AFLP data to test for topological incongruences.
Online Resource 5. Maximum credibility trees of post-burnin Bayesian analyses, each ran for 30 million generations, based on a. AFLP, b. nuclear (agt1, ETS, phyC) or c. plastid (atpB–rbcL, trnL–trnF, matK, ycf1 pos. 1114-2102 and 4550-5532) data.
Online Resource 6. Transition model selection for stochastic character mapping and state frequencies per node for Fig. 2. a. The corrected Akaike information criterion (AICc) was used for model selection. b. Table showing the inferred state frequencies calculated across 1000 stochastically mapped character histories (using ER model) for each node in Fig. 2 for two morphological traits: Apical coma (conspicuous, inconspicuous, missing) and Scape bracts (foliaceous, not foliaceous/often imbricate, lacking).
Online Resource 7. Overview of the published chromosome numbers in the genus Ananas and Pseudananas.
Rights and permissions
About this article
Cite this article
Matuszak-Renger, S., Paule, J., Heller, S. et al. Phylogenetic relationships among Ananas and related taxa (Bromelioideae, Bromeliaceae) based on nuclear, plastid and AFLP data. Plant Syst Evol 304, 841–851 (2018). https://doi.org/10.1007/s00606-018-1514-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-018-1514-3