Abstract
We studied the diversity, composition, and long-term dynamics of wood-inhabiting fungi in Quercus robur stumps left after commercial tree harvesting in Lithuania. Sampling of wood was carried out at three sites and from stumps, which were 10-, 20-, 30-, 40-, and 50-year-old. DNA was isolated from wood samples and fungal communities analyzed using high-throughput sequencing. Results showed that stump age had a limited effect on fungal diversity. The development of fungal communities in oak stums was found to be a slow process as fungal communities remained similar for decades, while larger changes were only detected in older stumps. The most common fungi were Eupezizella sp. (18.4%), Hyphodontia pallidula (12.9%), Mycena galericulata (8.3%), and Lenzites betulinus (7.1%). Fistulina hepatica, which is a red-listed wood-decay oak fungus, was also detected at a low relative abundance in stump wood. In the shortage of suitable substrate, oak stumps may provide habitats for long-term survival of different fungal species, including red-listed and oak-related fungi.
Similar content being viewed by others
Introduction
In Europe, common oak (Quercus robur) is a noble hardwood tree species, which is of key importance for forestry and nature conservation as it provides both valuable timber and habitats to a vast diversity of associated species [1, 2]. Oaks naturally remain viable for more than 300 years, but under optimal conditions some individuals can live for up to 1000 years or even longer [3]. During this long period of time, many different habitats are gradually formed in different parts of oak trees. For example, a coarse and rough bark starts to appear on trees of ca. 100-year-old, creating favorable conditions for the establishment of various lichen or moss species [4]. Such bark may also provide refuge for various insect species. In oaks of ca. 200-year-old, dead wood begins to appear in crowns, and such habitats only increase with tree age [5]. This creates conditions for the establishment of wood-decay fungi and wood-inhabiting insects. Lignin-decomposing fungi of the phylum Basidiomycota usually prevail in decaying oak wood and the activity of these creates conditions for the establishment of other fungi [6]. Changes in fungal species associated with dead wood are usually a slow process as these species are often long-lived, territorial, colonizing suitable areas, thus protecting themselves from other competing species [7]. As oaks age, the activity of wood-decay fungi and wood-inhabiting insects results in the appearance of decomposed wood, which also creates conditions for the establishment of larger organisms such as small mammals (e.g., dormouse, bats) or birds [8]. The latter observations show that for purposes of nature conservation the importance of oaks increases with the tree age.
According to some estimates, oaks could host nearly 900 different species [9, 10]. For example, Brändle and Brandl [11] showed that 252 phytophagous insect and mite species are oak specialists. Besides, the understory of oak-dominated woodlands is often characterized by a high diversity of plant species and soil-dwelling fungi [1]. In the Nordic countries, about 770 oak-associated species are nationally red-listed, about 400 of these are wood-inhabiting, of which about 300 are beetles and 50 are fungi [12]. Oaks host a higher number of red-listed species than any other tree species. Despite the importance, old-growth oaks have decreased not only in forests, but also in the agricultural landscape. They are often found in small populations or only as single trees [13]. Consequently, the ongoing decline of old-growth oaks has negative consequences for many associated species, especially many endangered species, which are often dependent on old oak habitats [14, 15].
Harvesting of oaks for wood is important for forestry and wood industry, but this also results in many oak stumps remaining in the forest for many decades, thereby providing a substrate for colonization. Due to the shortage of dead wood, and especially wood of larger dimensions, decaying oak stumps can be an important substrate for different saproxylic organisms [16]. The longevity of oak stumps, and oak wood in general, is due to the slow decay process, which is limited due to the presence of organic toxic compounds in the heartwood [17]. Following tree harvesting, the exposed stump surface is colonized by fungi through spores, followed by a gradual colonization of deeper wood layers and roots [18]. Some fungi colonizing stumps may also be present before the tree harvesting, including wood endophytes and latent invaders [7]. The emergence of fungi in stumps can also be linked to the presence of a nearby fungal colony or mycelial hyphal networks [19]. Over time, stumps become more accessible to these fungal species that simultaneously or selectively degrade lignin or modify lignin by degrading cellulose and hemicellulose [17, 20, 21], making such substrate more available to other organisms. In the past, understanding the diversity and composition of wood-inhabiting fungal communities was often problematic due to complexity of such communities and limitations of culture-based methods. Although the appearance of fungal fruiting bodies indicates the presence of one or another fungal species, this usually reflects only a fraction of entire fungal diversity. Besides, some fungi may be rare, but these may have a high conservation value [22,23,24]. The development of high-throughput sequencing methods allows the detection of numerous fungal species directly from environmental samples. These methods also provide semiquantitative information and a high resolution at the DNA sequence level.
The aim of this work was to study the diversity, composition, and long-term dynamics of wood-inhabiting fungi in oak stumps left after commercial tree harvesting in Lithuania. We hypothesized that (a) in decaying stumps, fungal diversity and community composition changes over time; (b) decaying stumps provide suitable habitats to fungi of conservation interest. In Lithuania, in the sixteenth century oak stands constituted between 15 and 20% of all forest stands, showing appropriate environmental conditions for oak to grow in this geographical area. Nowadays, however, oak stands constitute only ca. 2.0% of all forest stands and on average are 80-year-old. Old-growth oak stands are relatively rare, but several red-listed fungal species were reported in association with dead wood, including Ascomycetes such as Bactospora dryina, Calicium adspersum, Calicium quercinum, Cyphelium inquinans, Cladonia parasitica, Chaenotheca hispidula, Sclerophora coniophaea, and Basidiomycetes such as Fistulina hepatica, Hygrophorus russula, Inonotus dryophilus, Grifola frondosa, Piptoporus quercinus, Hapalopilus croceus, and Perenniporia medulla-panis.
Methods
Study Site and Sampling
The study sites were at Kėdainiai (K), Prienai (P), and Telšiai (T), which were in three different regions of Lithuania (Fig. 1). These regions are characterized by the highest density of Q. robur in the forest stands. At each site, Q. robur stumps were left after commercial clear-cuts and were between 10- and 50-year-old (Fig. 2). The age of stumps was determined based on available records of tree fellings. In each region, the distances between stumps of different age were up to 10 km. Woody vegetation varied in areas with stumps of different age, but often it was dominated by Q. robur with admixture of Picea abies, Betula pendula, Populus tremula, Tilia cordata, and Fraxinus excelsior. Understory consisted of Corylus avellana and Sorbus aucuparia. In different geographical regions, the composition of woody vegetation in vicinity to stumps of similar age was similar. Woody vegetation in vicinity of 10-year-old stumps was sparce.
Sampling of oak wood was carried out in 2021. At each site, five random stumps of each age, i.e., ca. 10-, 20-, 30-, 40-, and 50-year-old, were sampled (Fig. 2). Before the sampling, the 3-cm-thick wood layer was removed using a chainsaw, and three random wood cores (ca. 8-cm-long) per stump were taken using an increment borer (Haglöf, Sweden).
Wood samples collected from stumps of different age are shown in Fig. 3. Tools used for wood sampling were carefully cleaned between individual stumps. In total, there were 75 oak stumps sampled (3 regions × 5 age classes × 5 stumps). Collected wood samples were individually placed in sterile plastic tubes, labeled, the same day transported to the laboratory, and stored at −20 °C before further processing.
DNA Isolation, Amplification, and Sequencing
Three replicate wood cores from the same stump were put and analyzed together. The DNA work followed the study by Marčiulynas et al. [25]. Firstly, wood samples were ground in liquid nitrogen, and ca. 0.5 g of each stump wood sample was placed into a 2-mL screw-cap centrifugation tube together with two (2 mm in diameter) metal beads. Then, samples were homogenized using a Fast prep shaker (Montigny-le-Bretonneux, France), and the DNA was isolated using CTAB extraction buffer followed by incubation at 65 °C for 1 h. The supernatant was mixed with an equal volume of chloroform and cleaned with 2-propanol. The pellet was washed in 500 μL 70% ethanol, dried, and dissolved in 30 μL sterile milli-Q water. The DNA from each sample was further purified using a NucleoSpin®Soil kit (Macherey-Nagel GmbH & Co. Duren, Germany) according to the manufacturer’s recommendations. Following the isolation and purification of the DNA, the concentration was determined using a NanoDrop™ One spectrophotometer (Thermo Scientific, Rodchester, NY, USA) and DNA concentration adjusted to 10 ng/mL.
The amplification of the ITS2 rRNA region was achieved using a primer pair gITS7 [26] and ITS4 [27] both containing sample identification barcodes. The polymerase chain reaction (PCR) was performed in 50 μL reactions and consisted of the following final concentrations: 0.02 ng/μL template DNA, 200 μM dNTPs, 750 μM MgCl2, 0.025 μM DreamTaq Green polymerase (5 U/μL) (Thermo Scientific, Waltham, MA, USA), and 200 nM of each primer; sterile milli-Q water was added to make the final reaction volume of 50.0 μL. The amplifications were performed using an Applied Biosystems 2720 thermal cycler (Applied Biosystems, Foster City, CA, USA). The PCR program started with an initial denaturation step at 95 °C for 5 min, followed by 30 cycles of 95 °C for 30 s, and annealing at 55 °C for 30 s and 72 °C for 1 min, followed by a final extension step at 72 °C for 7 min. The PCR products were assessed using gel electrophoresis on 1.5% agarose gels stained with GelRed (Biotium, Fremont, CA, USA). The PCR products were purified using 3 M sodium acetate (pH 5.2) (Applichem GmbH, Darmstadt, Germany) and 96% ethanol mixture (1:20). After the quantification of all the PCR products using a Qubit fluorometer 4.0 (Life Technologies, Stockholm, Sweden), they were pooled in an equimolar mix and sequenced using a PacBio platform and one Sequel SMRT cell at a SciLifeLab facility in Uppsala, Sweden.
Bioinformatics
Generated sequences were subjected to quality control in the Sequence Clustering and Analysis of Tagged Amplicons (SCATA) bioinformatics pipeline (http://scata.mykopat.slu.se/ (accessed on 19 June 2023)). Sequences shorter than 200 pb, and sequences with low read quality (Q<20), and primer dimers were removed. Homopolymers were collapsed to 3 bp before clustering. Sequences that were missing a tag or primer were also removed. High-quality sequences were clustered into different OTUs using single linkage clustering based on 98.5% similarity. The GenBank (NCBI) database and Blastn algorithm were used to determine identities of different OTUs (Table S1). Taxon-delimiting ITS homology was 98–100% at the species level, 94–97% at the genus level, and 80–94% at the higher level, corresponding to at least 90% of the sequence length [28]. Sequences of fungal OTUs were deposited in GenBank under accession numbers OR164586-OR164872. FUNGuild was used to identify fungal functional groups (guilds) (FUNGuild v1.0) [29].
Statistical Analyses
Species accumulation curves were calculated using Analytical Rarefaction v.1.3, (http://www.uga.edu/strata/software/index.html). Differences in fungal OTU richness in Q. robur stumps of different age were compared using the non-parametric chi-square test [30]. The Shannon diversity index and Sørensen qualitative similarity index were calculated using SAS v. 9.4 (Cary, NC, USA) [30,31]. The non-parametric Mann-Whitney test in SAS was used to test whether Shannon's diversity index differed among stumps of different age. The composition of fungal communities was analyzed using non-metric multidimensional scaling (NMDS) based on the Bray-Curtis dissimilarity index. One-way analysis of similarity (ANOSIM) was performed to test for significant differences between different samples. A Principal Components Analysis (PCA) was performed to examine the impact of different sampling sites on fungal communities. These analyses were performed using Vegan 2.5.7 and Stats 3.6.2 in R 4.1.1 (https://www.r-project.org (accessed on 23 June 2023)) [32,33].
Results
High-throughput sequencing of fungal communities generated 179,782 reads, among which 69,127 were of high-quality and were retained, while 110,655 low quality reads were excluded. One sample had no data after quality filtering and was excluded (Telšiai, 20-year-old stump). Clustering analysis showed the presence of 350 non-singleton OTUs, among which 296 (84.6%) were fungal OTUs. Non-fungal OTUs and singletons were excluded. The number of high-quality sequences and fungal OTUs from stumps of different age and from different sampling sites is shown in Table 1. The Shannon diversity index of fungal communities was between 1.32 and 2.14 at the K site, between 0.92 and 1.78 at the P site, and between 1.23 and 2.26 at the T site (Fig. 4). The comparison of the Shannon diversity index of fungal communities in stumps of different age showed that in many cases these were similar and did not increase with the stump age (Fig. 4).
The Shannon diversity index of fungal communities in Quercus robur stumps of different age from three sampling sites (K—Kėdainiai, P—Prienai, and T—Telšiai) in Lithuania. Columns followed by the same letter do not differ significantly at p > 0.05. The comparison was done between fungal communities in stumps of different age classes and sampling sites
The plot of fungal OTUs vs. the number of fungal sequences resulted in species accumulation curves that did not reach the asymptote (Fig. 5). The OTU richness was similar among stumps of different age when the same number of sequences was taken per each stump age class (p > 0.05) (Fig. 5).
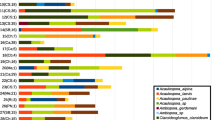
Among all fungal OTUs, 231 (78.0%) belonged to Ascomycota, 63 (21.3%) to Basidiomycota, and one to each (0.35%) Chytridiomycota and Mucoromycota. All fungal OTUs were found to belong to 18 different fungal classes, among which the most common were Leotiomycetes (28.9%), Sordariomycetes (17.4%) Agaricomycetes (13.2%), and Eurotiomycetes (13.2%) (Fig. 6).
The Sørensen qualitative similarity index of fungal communities in oak stumps of different age was low to moderate, ranging between 0.20 and 0.47 (Table 2).
The number of shared fungal OTUs between stumps of different age was lowest between 10- and 50-year-old stumps (Fig. 7). The number of unique fungal OTUs varied substantially among stumps of different age classes with the highest number found in 50-year-old stumps (Fig. 7). Overall, unique OTUs in stumps of different age constituted between 20.2% and 46.3% of all OTUs.
The fungal functional groups could be determined for 94.7% of fungal sequences, and their relative abundance in different sites and stumps of different age is shown in Fig. 8A. The analysis based on all sites, different stump age classes, and using fungal sequences showed that the most abundant fungal functional groups were undefined saprotrophs (32.3%), wood saprotrophs (31.4%), and endophytes (24.6%). Undefined saprotrophs showed the highest relative abundance in 30-year-old (55.5%) stumps and 50-year-old (50.8%) stumps. The highest relative abundance of wood saprotrophs was in 10-year-old (43.0%) stumps and 20-year-old (57.8%) stumps. The relative abundance of fungal endophytes was more pronounced in older oak stumps (Fig. 8A). The overall relative abundance of plant pathogenic fungi was 6.8%. However, in 10-year-old stumps, the relative abundance of plant pathogenic fungi was 22.2%, but it has significantly decreased in older stumps (p < 0.05) (Fig. 8A).
Relative abundance (%) of fungal functional groups in Quercus robur stumps of different age estimated based on fungal sequences (a) or on fungal OTUs (b). The numbers between diagrams represent the age of oak stumps. Other represent fungi, which are not associated with plants (e.g., animal pathogens)
The fungal functional groups could be determined for 61.7% of OTUs, and their relative abundance in different sites and stumps of different age is shown in Fig. 8B. Based on the OTUs data, the most abundant functional groups were undefined (38.3%), undefined saprotrophs (20.8%), and wood saprotrophs (12.4%). The relative abundance of other functional groups was below 10%. In stump of different age, the composition and relative abundance of different fungal functional groups were similar (p > 0.05) (Fig. 8B). Plant pathogenic fungi were found in all samples, but their relative abundance declined with the stump age, i.e., from 11.4% in 10-year-old stumps to 6.2% in 50-year-old stumps (Fig. 8B). By contrast, the relative abundance of wood saprotrophs has increased with the stump age, i.e., from 8.6% in 10-year-old stumps to 16.5% in 50-year-old stumps (Fig. 8B).
The 30 most frequently detected fungi from Q. robur stumps of different age, representing 92.2% of fungal sequences, is shown in Table 3. The most common OTUs were Eupezizella sp. 5630_5 (18.37%), Hyphodontia pallidula (12.9%), Mycena galericulata (8.3%), and L. betulinus (7.1%) (Table 3). The most common OTUs in 10-year-old stumps were L. betulinus (26.2%), Eupezizella sp. 5630_5 (12.2%), and Ascocoryne sarcoides (9.6%); in 20-year-old stumps, these were Hypoxylon rubiginosum (38.5%) and Laetiporus sulphureus (14.4%); in 30-year-old stumps—Megacollybia platyphylla (40.0%) and Armillaria cepistipes (13.7%); in 40-year-old stumps—Eupezizella sp. 5630_5 (40.7%) and Mycena galericulata (20.1%), and in 50-year-old stumps—Hyphodontia pallidula (45.3%) and Eupezizella sp. 5630_5 (22.6%) (Table 3). Among the 30 most common fungal OTUs, L. betulinus (26.2%), C. velutina (11.3%), Phlebia tremellosa (9.3%), Lentinus substrictus (3.9%), Peniophora incarnata (3.4%), and Unidentified sp. 5630_41 (1.4%) were unique in 10-year-old stumps. Similarly, M. platyphylla (40.0%) was unique in 30-year-old stumps, and H. parviporum (3.97%) and Phialocephala fusca (1.46%) in 50-year-old stumps, while there were no unique OTUs among the 30 most common OTUs in 20- and 40-year-old stumps (Table 3).
Among the 30 most common fungal OTUs and as compared among stumps of different age classes the relative abundance of Phacidiales sp. 5630_19 was significantly higher in 10-year-old stumps of H. rubiginosum—in 20-year-old stumps of Dasyscyphella sp. 5630_20, Eupezizella sp. 5630_22 and Mycena maculata—in 40-year-old stumps, and of H. pallidula—in 50-year-old stumps (p < 0.05).
Pathogenic and/or wood decay fungi such as Armillaria cepistipes (2.9%), L. sulphureus (2.7%), Phlebia tremellosa (2.5%), Stereum hirsutum (1.7%), H. parviporum (1.1%), and Peniophora incarnata (0.9%) were also detected among most common fungi (Table 3). Fistulina hepatica was the only red-listed wood decay oak fungus (Table S1), which was found at a low relative abundance in both 10-year-old stumps (0.06%) and in 50-year-old stumps (0.16%).
The NMDS showed a partial overlap of fungal communities in stumps that were 10-, 20-, 30-, and 40-year-old, but the lack of such overlap between fungal communities in stumps of 10- and 50-year-old (Fig. 9). The comparison showed that the composition of fungal communities differed significantly between stumps of 10-year-old and 30-year-old, 10-year-old and 40-year-old, and 10-year-old and 50-year-old (p < 0.05). A significant difference in the composition of fungal communities was also between stumps of 20-year-old and 50-year-old (p < 0.05), but not between stumps of other age (p > 0.05) (Fig. 9).
The PCA of fungal communities explained 17.3% of the variation on axis 1 and 14.2% on axis 2 (Fig. 10). Vectors of P and K sites pointed into a similar direction, while a vector of the T site pointed into an opposite direction, suggesting that the effect of different sites on fungal communities was either similar (P and K sites) or different (T site) (Fig. 10).
Discussion
The results demonstrated that the development of fungal communities in oak stumps is a slow process as these remained similar for several decades before a larger change could be observed (Fig. 9, Table 2). This may suggest that both stump age and environmental conditions present at each site can be important factors shaping these fungal communities (Fig. 10) as larger differences in this respect were between individual study sites (Fig. 8). It was shown that fungal communities colonizing living tree trunks persist in dead wood for years following tree wounding [34]. Some of the latent fungi already existing in living wood become primary colonizers immediately after the death of the tree [35]. Stumps can also be colonized by secondary fungi that enter through airborne spores and intensively decompose the lignocellulose [36, 37]. Stumps are eventually colonized by late colonizers such as soil cord forming fungi, which establish after secondary colonizers and complete during final stages of wood decay [38, 39]. During these stages of colonization and wood decay, the composition of fungal species often changes [40].
Although it is suggested that the abundance of fungal species in wood of oak stumps increases as the wood decay progresses [17], a similar trend was not yet observed in the present study. Consequently, the OTU diversity remailed generally similar in stumps of different age, demonstrating that there was little or no accumulation of fungal diversity over time (Figs. 4 and 7). Previous studies showed positive, neutral, or negative relationship between fungal species richness and wood decay, which is measured as a decrease in wood weight or density [41, 42]. Wood core samples from stumps of different age were also similar in their appearance (Fig. 3), suggesting that the process of wood decay was slow. This can be due to the fact that the relative abundance of wood saprotrophs remained largely unchanged in stumps of different age (Fig. 8). Only wood saprotrophs are able to degrade lignin and access cellulose, which together with hemicellulose is the main source of energy for wood-degrading fungi [43, 44]. The results may therefore suggest that the establishment and activity of wood saprotrophs were limited by the competitive capacity of indigenous fungal community and/or oak wood properties. The abundance of wood saprotrophs generally increases in the advanced stage of wood decay [45, 46]. Parisi et al. [47], but the results of the present study indicate that more than 50 years may be required to reach such stage of decay in oak stumps.
Nevertheless, certain differences in the distribution of different saprotrophic species were in stumps of different age (Table 3). For example, L. betulinus and Phlebia tremellosa were found only in 10-year-old stumps, indicating that these fungi are the primary colonizers of dead stump wood. L. betulinus was shown to be one of the primary wood colonizers of living Quercus castaneifolia trees [48]. P. tremellosa is a lignin-degrading wood-decay fungus, which apart from wood of hardwoods, can also colonize weakened but still living oaks [49, 50]. Stereum hirsutum was present in 10- and 20-year-old stumps, which is in agreement with the previous observation that it is an early colonizer of oak wood [34]. H. rubiginosum and L. sulphureus were wood-decay fungi, which were primarily found in 20- and 30-year-old stumps (Table 3). [49] suggested that H. rubiginosum is a pioneer fungal species, which grows in living or recently dead trees, causing a slow decay. H. rubiginosum was also found in oak stems characterized by late stage of decay [51]. The latter observations may suggest that H. rubiginosum is able to establish in fresh wood and continue wood decomposition for many years. L. sulphureus is known to cause heart rot in living trees, and after the death of the tree, it continues to decompose dead standing or fallen stems and stumps [52, 53]. L. sulphureus was also reported from highly decomposed oak wood [52]. Interestingly, H. parviporum, which is a primary pathogen and wood decay fungus of Picea abies in northern Europe [54, 55], was detected in 50-year-old stumps, suggesting that in oak wood it can be a secondary colonizer.
Plant-pathogenic fungi were most abundant in 10-year-old stumps (Fig. 8A), but in older stumps their abundance declined even though the proportion of their OTUs remained largely unchanged over time (Fig. 8B). Commonly detected pathogenic fungi included Armillaria cepistipes and H. parviporum (Table 3), which can often be found in the lower part of tree trunks and/or in the roots [56, 57]. Such colonized substrate can be the source of disease infection to the neighboring trees through root contact [58]. Diplodia mutila was also a commonly detected pathogen, which was shown to be associated with cankers on Q. petraea [59] and Q. robur [60]. Another commonly detected pathogen was Ophiostoma quercus, which is found on Quercus and Fagus trees in the northern hemisphere [61] and is often associated with oak decline in Central and Eastern Europe [62].
The study also showed that oak stumps can be a suitable habitat for protected and red-listed species such as a wood-decay fungus F. hepatica (Table S1). F. hepatica is usually found in still-growing oak trees and is known to cause a condition known as brown oak [63], in which reddish brown spots appear on the heartwood of oaks during the early stages of fungal development [64]. F. hepatica also contributes to the formation of oak hollows, thereby forming principal habitats for endangered beetles. The presence of F. hepatica in both 10-year-old and 50-year-old stumps (Table S1) showed that due to the long-lasting decay of oak stumps, suitable conditions for the survival of such fungi are maintained for a long time.
Conclusions
Decaying oak stumps provide habitats to a high diversity of fungal species including red-listed and oak-related fungi, thereby supporting biodiversity. In the shortage of suitable substrate, oak stumps may create conditions for long-term survival of different fungal species. The development of fungal communities in oak stums is a gradual and slow process as fungal communities established in oak wood remain similar for decades.
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information file.
References
Leuschner C, Ellenberg H (2017) Ecology of Central European non-forest vegetation: coastal to alpine, natural to man-made habitats: vegetation ecology of Central Europe, vol 2. Springer II, p 1094. https://doi.org/10.1007/978-3-319-43048-5
Mölder A, Meyer P, Nagel RV (2019) Integrative management to sustain biodiversity and ecological continuity in Central European temperate oak (Quercus robur, Q. petraea) forests: an overview. For Ecol Manag 437:324–339. https://doi.org/10.1016/j.foreco.2019.01.006
Farjon A (2022) Ancient oaks in the English landscape. Kew Publishing, Kew, UK, p 352
Nordén B, Götmark F, Tönnberg M, Ryberg M (2004) Dead wood in semi-natural temperate broadleaved woodland: contribution of coarse and fine dead wood, attached dead wood and stumps. For Ecol Manag 194(1-3):235–248. https://doi.org/10.1016/j.foreco.2004.02.043
Dahlberg A, Croneborg H (2004) The 33 threatened fungi in Europe. Council of Europe. Mycol Res 108(1). https://doi.org/10.1017/S0953756204259287
Rytioja J, Hilden K, Yuzon J (2014) Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiol Mol Biol R78:614–649
Boddy L, Heilmann-Clausen J (2008) Basidiomycete community development in temperate angiosperm wood. British Mycological Society Symposia Series, vol 28. Academic Press, pp 211–237. https://doi.org/10.1016/S0275-0287(08)80014-8
Kõrkjas M, Remm L, Lõhmus A (2021) Development rates and persistence of the microhabitats initiated by disease and injuries in live trees: a review. For Ecol Manag 482:118833. https://doi.org/10.1016/j.foreco.2020.118833
Jansson N, Bergman KO, Jonsell M, Milberg P (2009) An indicator system for identification of sites of high conservation value for saproxylic oak (Quercus spp.) beetles in southern Sweden. J Insect Conserv 13:399–412. https://doi.org/10.1007/s10841-008-9187-9
Sundberg S, Carlberg T, Sandström J, Thor G (2019) Värdväxters betydelse för andra organismer–med fokus på vedartade värdväxter. SLU Artdatabanken, Sweden, p 52
Brändle M, Brandl R (2001) Species richness of insects and mites on trees: expanding Southwood. J Anim Ecol 70(3):491–504. https://doi.org/10.1046/j.1365-2656.2001.00506.x
Tingstad L, Grytnes JA, Felde VA, Juslén A, Hyvärinen E, Dahlberg A (2018) The potential to use documentation in national Red Lists to characterize red-listed forest species in Fennoscandia and to guide conservation. GECCO 15:e00410. https://doi.org/10.1016/j.gecco.2018.e00410
Peterken GF (1996) Natural woodland: ecology and conservation in northern temperate regions. Cambridge university press, Cembridge, UK, p 522
Sunhede S, Vasiliauskas R (1996) Wood and bark inhabiting fungi on oak in Lithuania. Balt For 2:23–27
Sunhede S, Vasiliauskas R (2003) Ecology and decay pattern of Inocutis dryophila on Quercus robur. Karstenia 43(2):45–53. https://doi.org/10.29203/ka.2002.380
Steiner KC (1998) A decline-model interpretation of genetic and habitat structure in oak populations and its implications for silviculture. Eur J For Path 28(2):113–120. https://doi.org/10.1111/j.1439-0329.1998.tb01172.x
van der Wal A, Ottosson E, De Boer W (2015) Neglected role of fungal community composition in explaining variation in wood decay rates. Ecol 96(1):124–133. https://doi.org/10.1890/14-0242.1
Pearce MH, Malajczuk N (1990) Stump colonization by Armillaria luteobubalina and other wood decay fungi in an age series of cut-over stumps in karri (Eucalyptus diversicolor) regrowth forests in south-western Australia. New Phytol 115(1):129–138. https://doi.org/10.1111/j.1469-8137.1990.tb00930.x
Hiscox J, Boddy L (2017) Armed and dangerous–chemical warfare in wood decay communities. Fungal Biol Rev 31(4):169–184. https://doi.org/10.1016/j.fbr.2017.07.001
Schilling JS, Kaffenberger JT, Held BW, Ortiz R, Blanchette RA (2020) Using wood rot phenotypes to illuminate the “gray” among decomposer fungi. Front Microbiol 11:1288. https://doi.org/10.3389/fmicb.2020.01288
Schilling JS, Kaffenberger JT, Liew FJ, Song Z (2015) Signature wood modifications reveal decomposer community history. PLoS One 10:e0120679. https://doi.org/10.1371/journal.pone.0126877
Berglund H, Jönsson MT, Penttilä R, Vanha-Majamaa I (2011) The effects of burning and dead-wood creation on the diversity of pioneer wood-inhabiting fungi in managed boreal spruce forests. For Ecol Manag 261(7):1293–1305. https://doi.org/10.1016/j.foreco.2011.01.008
Nordén J, Abrego N, Boddy L, Bässler C, Dahlberg A, Halme P, Hällfors M, Maurice S, Menkis A, Miettinen O, Mäkipää R, Ovaskainen O, Penttilä R, Saine S, Snäll T, Junninen K (2020) Ten principles for conservation translocations of threatened wood-inhabiting fungi. Fungal Ecol 44:100919. https://doi.org/10.1016/j.funeco.2020.100919
Toivanen T, Markkanen A, Kotiaho JS, Halme P (2012) The effect of forest fuel harvesting on the fungal diversity of clear-cuts. Biomass Bioenergy 39:84–93. https://doi.org/10.1016/j.biombioe.2011.11.016
Marčiulynas A, Marčiulynienė D, Lynikienė J, Bakys R, Menkis A (2022) Fungal communities in leaves and roots of healthy-looking and diseased Ulmus glabra. Microorganisms 10:2228. https://doi.org/10.3390/microorganisms10112228
Ihrmark K, Bodeker ITM, Cruz-Martinez K, Friberg H, Kubartova A, Schenck J, Strid Y, Stenlid J, Brandstrom-Durling M, Clemmensen KE (2012) New primers to amplify the fungal ITS2—evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol 82:666–677. https://doi.org/10.1111/j.1574-6941.2012.01437.x
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA, USA, pp 315–322
Kubartová A, Ottosson E, Dahlberg A, Stenlid J (2012) Patterns of fungal communities among and within decaying logs, revealed by 454 sequencing. Mol Ecol 21(18):4514–4532. https://doi.org/10.1111/j.1365-294X.2012.05723.x
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) FUNGuild: an OpenAnnotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248. https://doi.org/10.1016/j.funeco.2015.06.006
Magurran AE (1988) Ecological diversity and its measurement. Princeton University Press, Princeton, NJ, USA, p 192
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:379–423
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. Community ecology package. R package version 2. (2013) Available online: https://cran.r-project.org/web/packages/vegan/index.html. Accessed 23 June 2023
R Core Team (2021) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Menkis A, Redr D, Bengtsson V, Hedin J, Niklasson M, Nordén B, Dahlberg A (2022) Endophytes dominate fungal communities in six-year-old veteranisation wounds in living oak trunks. Funct Ecol 59:101020. https://doi.org/10.1016/j.funeco.2020.101020
Parfitt D, Hunt J, Dockrell D, Rogers HJ, Boddy L (2010) Do all trees carry the seeds of their own destruction? PCR reveals numerous wood decay fungi latently present in sapwood of a wide range of angiosperm trees. Fungal Ecol 3(4):338–346. https://doi.org/10.1016/j.funeco.2010.02.001
Coates D, Rayner ADM (1985) Fungal population and community development in cut beech logs: I. Establishment via the aerial cut surface. New Phytol 101(1):153–171. https://doi.org/10.1111/j.1469-8137.1985.tb02824.x
Fukasawa Y, Osono T, Takeda H (2011) Wood decomposing abilities of diverse lignicolous fungi on nondecayed and decayed beech wood. Mycologia 103(3):474–482. https://doi.org/10.3852/10-246
Boddy L (2001) Fungal community ecology and wood decomposition processes in angiosperms: from standing tree to complete decay of coarse woody debris. Ecol Bull:43–56 http://www.jstor.org/stable/20113263
Fukasawa Y (2021) Ecological impacts of fungal wood decay types: a review of current knowledge and future research directions. Ecol Res 36(6):910–931. https://doi.org/10.1111/1440-1703.12260
Boddy L, Rayner ADM (1983) Mycelial interactions, morphogenesis and ecology of Phlebia radiata and P. rufa from oak. TBMS 80(3):437–448. https://doi.org/10.1016/S0007-1536(83)80040-0
Dickie IA, Fukami T, Wilkie JP, Allen RB, Buchanan PK (2012) Do assembly history effects attenuate from species to ecosystem properties? A field test with wood-inhabiting fungi. Ecol Lett 15(2):133–141. https://doi.org/10.1111/j.1461-0248.2011.01722.x
Purahong W, Wubet T, Krüger D, Buscot F (2018) Molecular evidence strongly supports deadwood-inhabiting fungi exhibiting unexpected tree species preferences in temperate forests. The ISME J 12(1):289–295. https://doi.org/10.1038/ismej.2017.177
Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, Asiegbu FO, Baker SE, Barry K, Bendiksby M, Blumentritt M, Coutinho PM, Cullen D, de Vries RP, Gathman A, Goodell B, Henrissat B, Ihrmark K, Kauserud H et al (2011) The plant cell wall–decomposing machinery underlies the functional diversity of forest fungi. Science 333(6043):762–765. https://doi.org/10.1126/science.1205411
Martínez AT, Rencoret J, Nieto L, Jiménez-Barbero J, Gutiérrez A, Del Río JC (2011) Selective lignin and polysaccharide removal in natural fungal decay of wood as evidenced by in situ structural analyses. Environ Microbiol 13(1):96–107. https://doi.org/10.1111/j.1462-2920.2010.02312
Ottosson E, Norden J, Dahlberg A, Edman M, Jönsson M, Larsson KH, Olsson J, Penttilä R, Ovaskainen O (2014) Species associations during the succession of wood-inhabiting fungal communities. Fungal Ecol 11:17–28. https://doi.org/10.1016/j.funeco.2014.03.003
Rajala T, Tuomivirta T, Pennanen T, Mäkipää R (2015) Habitat models of wood-inhabiting fungi along a decay gradient of Norway spruce logs. Fungal Ecol 18:48–55. https://doi.org/10.1016/j.funeco.2015.08.007
Parisi F, Pioli S, Lombardi F, Fravolini G, Marchetti M, Tognetti R (2018) Linking deadwood traits with saproxylic invertebrates and fungi in European forests-a review. iForest 11(3):423. https://doi.org/10.3832/ifor2670-011
Bari E, Karimi K, Aghajani H, Schmidt O, Zaheri S, Tajick-Ghanbary MA, Juybari HZ (2021) Characterizations of tree-decay fungi by molecular and morphological investigationsin aniranian alamdardeh forest. Maderas-Cienc Technol 23. https://doi.org/10.4067/s0718-221x2021000100433
Rayner ADM, Boddy L (1988) Fungal communities in the decay of wood. Adv Microb Ecol 10:115–166. https://doi.org/10.1007/978-1-4684-5409-3_4
Rayner ADM, Boddy L (1989) Fungal decomposition of wood. Its biology and ecology. Forest Sci 35(2):647–648. https://doi.org/10.1093/forestscience/35.2.647
Iršėnaitė R, Kutorga E (2006) Diversity of fungi on decaying common oak coarse woody debris. Ekologija 4:22–30 http://elibrary.lt/resursai/LMA/Ekologija/Eko64/Eko64_07.pdf
Ryvarden L, Gilbertson RL (1993) European polypores: Part 1: Abortiporus-Lindtneria. Department of Botany, University of Oslo, Blindern, Oslo, Norway, p 387
Vasaitis R, Menkis A, Lim YW, Seok S, Tomsovsky M, Jankovsky L, Lygis V, Slippers B, Stenlid J (2009) Genetic variation and relationships in Laetiporus sulphureus s. lat., as determined by ITS rDNA sequences and in vitro growth rate. Mycol Res 113(3):326–336. https://doi.org/10.1016/j.mycres.2008.11.009
Arnerup J, Lind M, Olson Å, Stenlid J, Elfstrand M (2011) The pathogenic white-rot fungus Heterobasidion parviporum triggers non-specific defence responses in the bark of Norway spruce. Tree Physiol 31(11):1262–1272. https://doi.org/10.1093/treephys/tpr113
Szczepkowski A, Kowalczuk W, Sikora K, Damszel M, Sierota Z (2022) Fungi occurring in Norway spruce wood decayed by Heterobasidion parviporum in Puszcza Borecka Stands (northeastern Poland). Forests 13:229. https://doi.org/10.3390/f13020229
Cleary M, van der Kamp B, Morrison D (2008) British Columbia’s southern interior forests Armillaria root disease stand establishment decision aid. JEM 9(2):60–65. https://doi.org/10.22230/jem.2008v9n2a397
Oliva J, Bendz-Hellgren M, Stenlid J (2011) Spread of Heterobasidion annosum ss and Heterobasidion parviporum in Picea abies 15 years after stump inoculation. FEMS Microb Ecol 75(3):414–429. https://doi.org/10.1111/j.1574-6941.2010.01020.x
Bendz-Hellgren M, Stenlid J (1998) Effects of clear-cutting, thinning, and wood moisture content on the susceptibility of Norway spruce stumps to Heterobasidion annosum. Can J For Res 28(5):759–765. https://doi.org/10.1139/x98-043
Vajna L (1986) Branch canker and dieback of sessile oak (Quercus petraea) in Hungary caused by Diplodia mutila. I. Identification of the pathogen. Eur J For Pathol 16(4):223–229 https://www.cabdirect.org/cabdirect/abstract/19861319603
Moricca S, Ragazzi A (2008) Fungal endophytes in Mediterranean oak forests: a lesson from Discula quercina. Phytopathology 98(4):380–386. https://doi.org/10.1094/PHYTO-98-4-0380
Brasier CM, Kirk SA (1993) Sibling species within Ophiostoma piceae. Mycol Res 97(7):811–816. https://doi.org/10.1016/S0953-7562(09)81156-8
Macháčová M, Nakládal O, Samek M, Baťa D, Zumr V, Pešková V (2022) Oak decline caused by biotic and abiotic factors in Central Europe: a case study from the Czech Republic. Forests 13:1223. https://doi.org/10.3390/f13081223
Cartwright KTSG (1937) A reinvestigation into the cause of ‘Brown Oak’, Fistulina hepatica (Hus.) Fr. Trans Br Mycol Soc 40:17–89
Schwarze FWMR (2004) Forest Pathology: Heart Rot and Wood Decay. In: Burley JJ, Evans J. Youngquist (eds) Encylopedia of Forest Sciences, Elsevier Science, pp 808–816. https://enviro2.doe.gov.my/ekmc/wp-content/uploads/2016/08/1384851791-3-s2.0-B0121451607000715-main.pdf
Funding
This work was supported by the European Social Fund under the 09.3.3-LMT-K-712 “Development of Competences of Scientists other Researchers and Students through Practical Research Activities” measure.
Author information
Authors and Affiliations
Contributions
A.M. and A.M. designed the experiments; A.M. carried out the experiments, analyzed the data, and wrote the manuscript; A.M. reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Supplementary Information
ESM 1
(XLSX 57 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Marčiulynas, A., Menkis, A. Long-term Dynamics of Fungal Communities Inhabiting Decaying Stumps of Quercus robur. Microb Ecol 87, 27 (2024). https://doi.org/10.1007/s00248-023-02334-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00248-023-02334-3